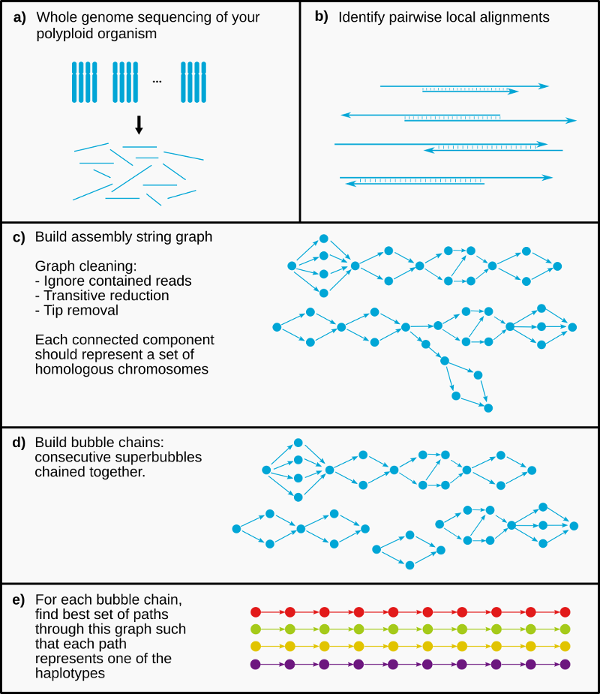
PHASM is a long read de novo genome assembler that phases variants among chromosome homologues during the assembly process, and aims to output separate contigs for each haplotype. The main idea in PHASM is to build bubble chains: consecutive “superbubbles” chained together. While most traditional genome assemblers pop these superbubbles by only keeping the best supported path, PHASM finds k paths through this chain of superbubbles that best represent each haplotype.
This program has been created as part of my master thesis project. For now, it has only been tested with error free data.
- Status: Finished.
- License: MIT
Overview
Pipeline
The PHASM pipeline consists of four main stages:
- Overlapper
- Assembly graph construction
- Bubblechain identification
- Phasing
Key Points
- Main idea:
- Different alleles on homologous chromosomes result in branches in the assembly graph;
- A bubble or superbubble is a common motif in a genome assembly graph;
- Hypothesis: each superbubble has a valid path from its source to its sink that spells a DNA sequence that matches with one of the haplotypes. Furthermore, we expect that for each haplotype, there exists at least one path through this superbubble that spells a matching DNA sequence.
- A lot of current genome assemblers pop bubbles in the assembly graph, thereby removing variation among homologous chromosomes
- Build an assembler that keeps superbubbles and searches for the best set of paths such that each path represents a haplotype.
- Main findings:
- Superbubbles are an incomplete representation of variation among homologous chromosomes
- Paths through a superbubble are rarely consistent with one of the haplotypes
- Superbubbles are probably an artefact of approximate overlapping between reads.
Read more in my master thesis.
Availability
PHASM is available on Github.