Urinary tract infections (UTIs) are common, painful, and a large burden on healthcare worldwide. They frequently become recurrent, with some getting an infection multiple times a year. Furthermore, treatment becomes more challenging with increasing antibiotic resistance among the bacteria that cause UTIs. It is thus important to understand what drives UTI recurrence and devise treatment strategies that do not require antibiotics.
Escherichia coli is the most common bacterial species causing UTIs, and the gut is a known reservoir of uropathogenic (UPEC) strains. Less understood, however, is the role of the entire gut microbiome in facilitating recurrence and whether any immunological differences play a role.
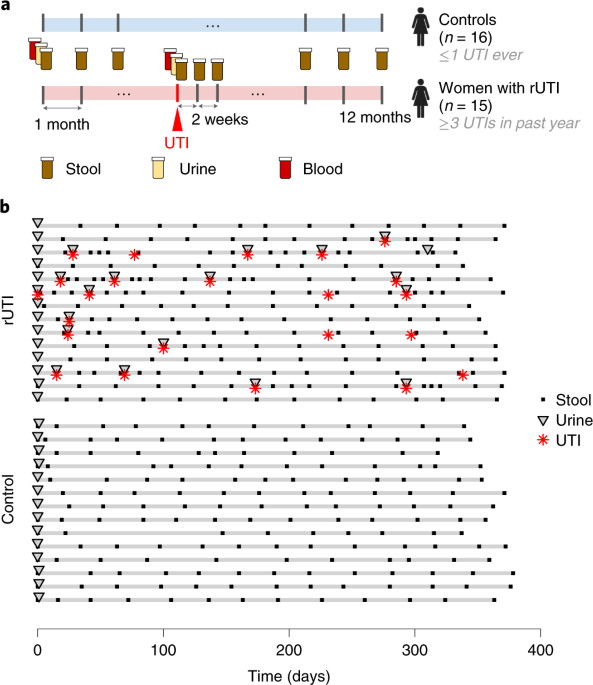
In this paper, we studied women with a history of recurrent UTIs for a year, collecting monthly blood, stool, and urine samples, and additional samples when diagnosed with a UTI. This enabled us to characterize the dynamics of the gut microbiome and explore its relationship with recurrent UTIs. Blood samples additionally provided a snapshot of their immunological state. We compared women in our UTI cohort with a matched cohort of healthy women.
Our main findings include (1) the gut microbiome of women with UTIs are significantly less diverse, with fewer bacterial species present than typically seen in healthy women and showing more characteristics of low-level inflammation, (2) immunological biomarkers suggested a distinct immunological state, (3) E. coli strains frequently transmitted between the gut and the bladder in both the healthy cohort and the UTI cohort, though healthy women did not exhibit any UTI symptoms, and (4) the UTI-causing strain was rarely cleared from the gut after antibiotic treatment.
The latter two findings were enabled by StrainGE, a software tool we specifically designed to identify and characterize low-abundance strains in complex microbial communities. It can untangle same-species strain mixtures and identify strain-specific genetic variants, enabling the tracking of strains across samples.